Normalization of agilent Seahorse™ XF data with direct cell counting using the Celigo image cytometer
In recent years, the Seahorse XF Analyzer has been utilized to measure the Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR)/ Proton Production Rate (PPR) for understanding energy metabolism and cellular functions [1].
One of the important steps in performing Seahorse XF cell metabolism analysis is normalization of the resultant data to reduce inconsistency and variations well-to-well due to potential non-uniform cell seeding across the microplate. In general, normalization has been commonly performed using total protein amount in each well such as BCA assays [2].
There are some known issues with the protein-based normalization method:
- Cells can be lost during the assay due to the metabolic inhibitors used during the assay.
- Normalizing Seahorse data using total protein analysis is time-consuming. It requires multiple steps such as cell lysing, incubation, and transferring lysed content for analysis.
- The washing and transfer procedure can introduce risk such as cell loss or pipetting error, thus skewing the final normalized results.
- The BCA assay can take up to ~45 min for the entire 96-well plate.
An alternative method that can improve Seahorse XF normalization is to perform a direct cell count per well prior to the metabolic stress tests.
The direct cell count-based normalization method reduces issues from protein analysis
- Cell counting is a non-destructive method to generate data for Seahorse normalization
- It can take account of non-uniform seeding density across the well and plate
- Downstream assays are still possible after normalization
- Direct cell counting can significantly reduce the amount of time required to perform normalization
Using Celigo image cytometer for whole well direct cell counting
The Celigo™ image cytometer has been used to perform brightfield or fluorescence-based direct cell counting for Seahorse XF data normalization [3]. Due to its ability to capture whole-well images, at high speed, the Celigo direct cell counting application has been adapted into the general workflow for Seahorse XF96, XFe96, and XF24 Analyzers.
The Celigo image cytometer provides a fast, effective, and cell stress-free technique to count cells plated onto uncoated or coated culture plates. The system contains built-in plate profiles for the Seahorse plates and will acquire and analyze whole well images of an entire 96-well plate in ~6 min in brightfield, and ~9 min in fluorescence.
The Celigo image cytometer can be used to perform direct cell counting of suspension and adherent cells in brightfield, as well as fluorescence such as Hoechst 33342 or live-nuclear dye staining. Both brightfield and fluorescence methods can be used effectively to normalize Seahorse XF data.
The use of live nuclear dyes (Revvity) can result in a brighter fluorescence signal and reduced cytotoxic & cytokinetic effects. In addition, the cell identification algorithms are highly reliable for segmenting suspension or adherent cells for both brightfield and fluorescence imaging. Fluorescent live nuclear dyes can help improve accuracy for counting adherent cells that tend to flatten out after attachment.
Furthermore, the Celigo system has a large dynamic range of counting cells in whole well, i.e. counting 1 to ~120,000 cells per well for suspension cells and 1 to ~90,000 cells for adherent cells in a 96-well plate.

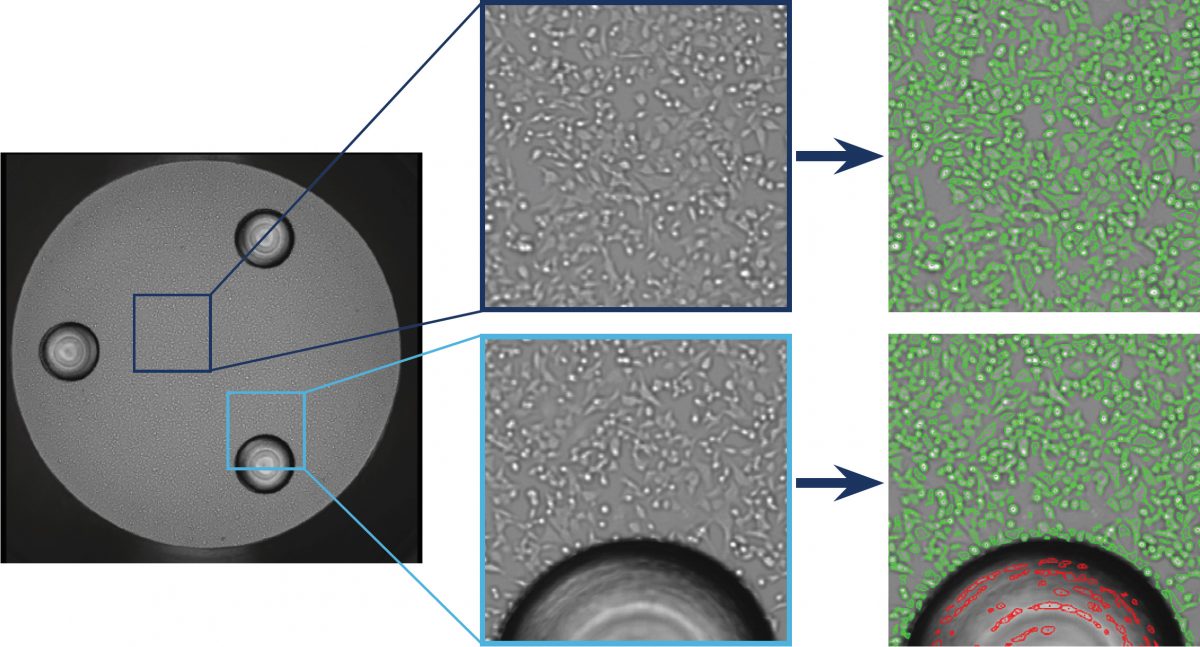
Figure 1. Whole-well images captured by the Celigo instrument from a Seahorse XF well with 3 posts. Direct cell count is shown where cells are circled in green and exclusion of the post is shown in red.
Brightfield-based direct cell counting method for Seahorse XF normalization
In this experiment, the glycolytic function (ECAR) was assessed for DCIS and HCT116 cell lines for approximately 160 min. The Seahorse XF data were normalized to both protein amount and brightfield cell counting using the Celigo image cytometer. The normalized time-dependent ECAR data were then compared, which showed improvement in cell line differentiation using the cell count normalization.
- DCIS and HCT116 cells were seeded at 12,500 cells/well and 10,000 cells/well, respectively, in the Seahorse XF 96-well plate and incubated overnight.
- The plate was first visualized on a microscope for approximately 95% confluence.
- The confluence was then confirmed at 98% using the Celigo system, as well as QC for cell health, morphology, seeding uniformity and purity (no contamination) prior to washing.
- Next, washed with the appropriate assay media.
- Subsequently, the plate was imaged and analyzed on the Celigo instrument using direct cell counting application.
- Next, the plate was placed in a 37° C incubator without CO2 for 45 minutes.
- The plate was then analyzed using Seahorse XFe96 to measure ECAR for 160 min.
- Finally, the plate was used to perform protein analysis for normalization.
The Celigo image cytometer-captured brightfield images are displayed in Figure 1, which show a whole well image of a Seahorse XF well with the 3 posts. In addition, the zoomed in images show direct cell counting of the HC116 cells (green outline), as well as exclusion of the post area (red outline).
By utilizing the graphing function and well mask feature in the Celigo software, researchers can count cells at specific percentages of area within the well, which may allow better normalization for the Seahorse XF data (Figure 2).
The normalized ECAR time-dependent data for protein and cell count normalization are shown in Figure 3. The data shows that measurements with protein normalization cannot distinguish between the glycolytic function of DCIS and HCT116 cell lines. In contrast, the cell count normalization method shows a reduction in standard deviation as well as a clear separation between the ECAR values of DCIS and HCT116 cell lines.
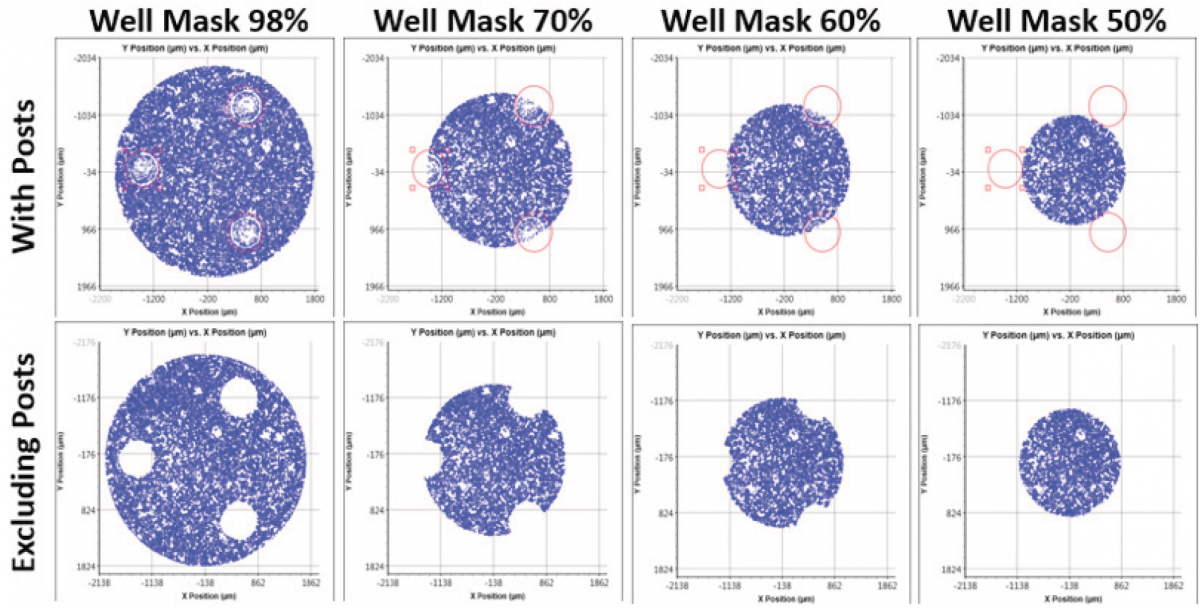
Figure 2. The graphing and well mask function of the Celigo software allows for cell counts at specific percentages within the area of a well.
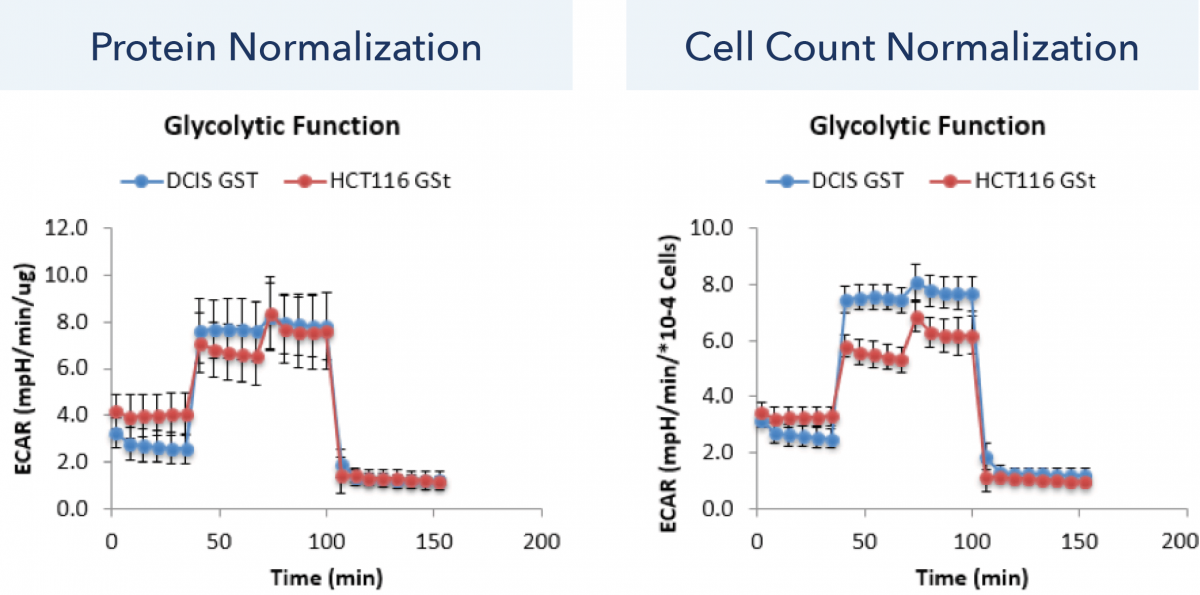
Figure 3. Normalized ECAR time-dependent data for protein and cell count normalization.
Fluorescence-based direct cell counting method for Seahorse XF normalization
In this experiment, both mitochondrial respiration (OCR) and glycolytic function (ECAR) were assessed for T cells for approximately 60 min. The Seahorse XF data were normalized to both protein amount and fluorescent cell counting using the Celigo Image Cytometer. The normalized endpoint ECAR data were then compared, which showed improvement in drug response differentiation using the cell count normalization.
- T cells were seeded at 100,000 cells/well in the Seahorse XF 96-well plate.
- The cells were allowed to settle to the bottom of the plate.
- The plate was then placed into the Seahorse XF Analyzer and allowed delivery of Hoechst 33342, and subsequently analyzed for 60 min.
- The plate was then counted on the Celigo for total Hoechst positive cells.
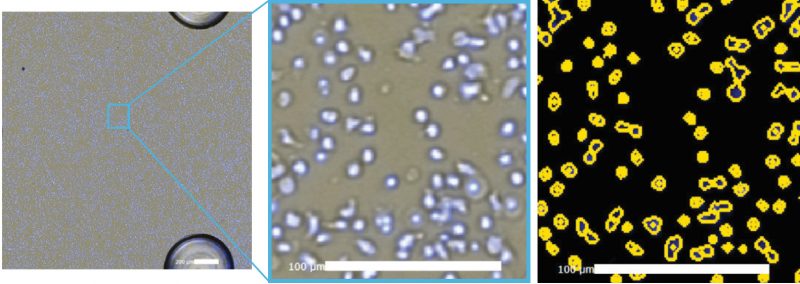
Figure 4. Celigo-captured brightfield image (left), zoomed merged brightfield and Hoechst image (center), and yellow ROI counted Hoechst-stained cells (right).
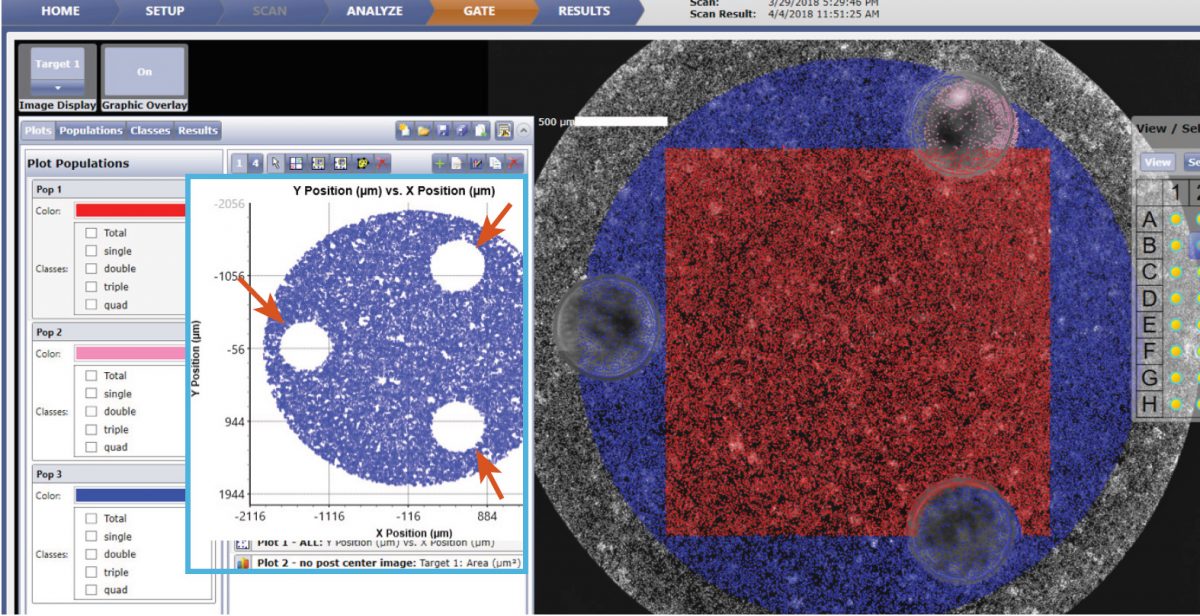
Figure 5. Using the Celigo gating function, the location graph is generated to gate out the 3 posts (indicated by red arrows above) in the well using fluorescence. In addition, if only the area between the 3 posts is analyzed (shown with red square), scanning and counting require only 2 min per plate.
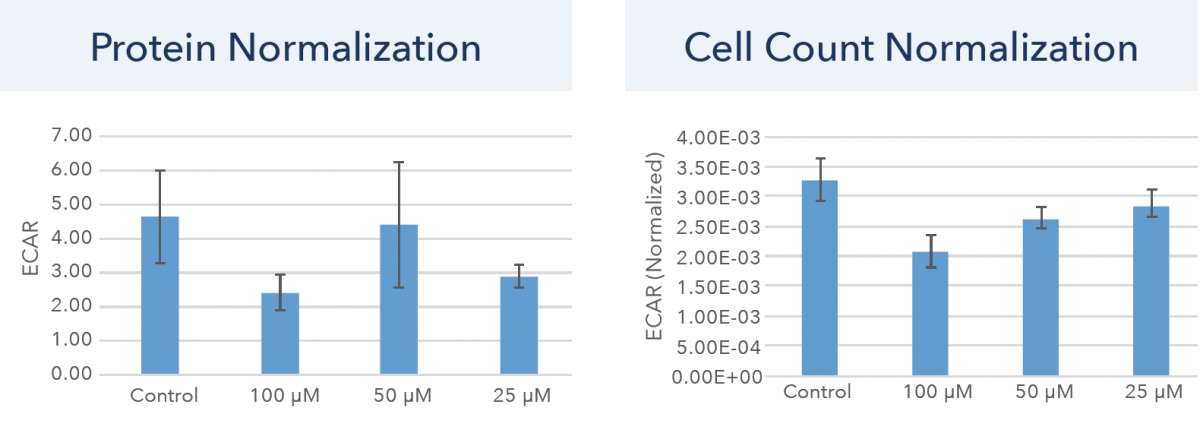
Figure 6. Normalized ECAR endpoint data for protein and cell count normalization.
The Celigo-captured brightfield and Hoechst fluorescence overlay images are shown in Figure 4. The counted image shows yellow outlines indicating counted Hoechst positive cells. Similarly, under the gating function, the location graph can be generated to gate out the 3 posts in the well using fluorescence (Figure 5).
The normalized ECAR endpoint data for protein and cell count normalization are shown in Figure 6. Similarly, the protein and cell count normalization method shows distinct results. Using protein normalization, a large standard deviation for each sample can be observed, as well as inexplicable drug treatment trends. On the other hand, the cell count normalization shows a reduction in standard deviation, as well as showing expected drug treatment results with a decrease in ECAR as drug concentration increases.
Speed to results
The Celigo image cytometer can scan whole-well images and directly count cells in less than 6 and 9 min per 96-well plate (less than 4 and 5 seconds per well) using brightfield and fluorescence imaging, respectively (Figure 7). If only the area between the 3 posts is captured, the entire process takes approximately 2 mins. Researchers investigating mitochondrial respiration and glycolytic functions using Seahorse XF Analyzer can rapidly and effectively normalize the OCR and ECAR measurements without additional procedures like protein analysis.
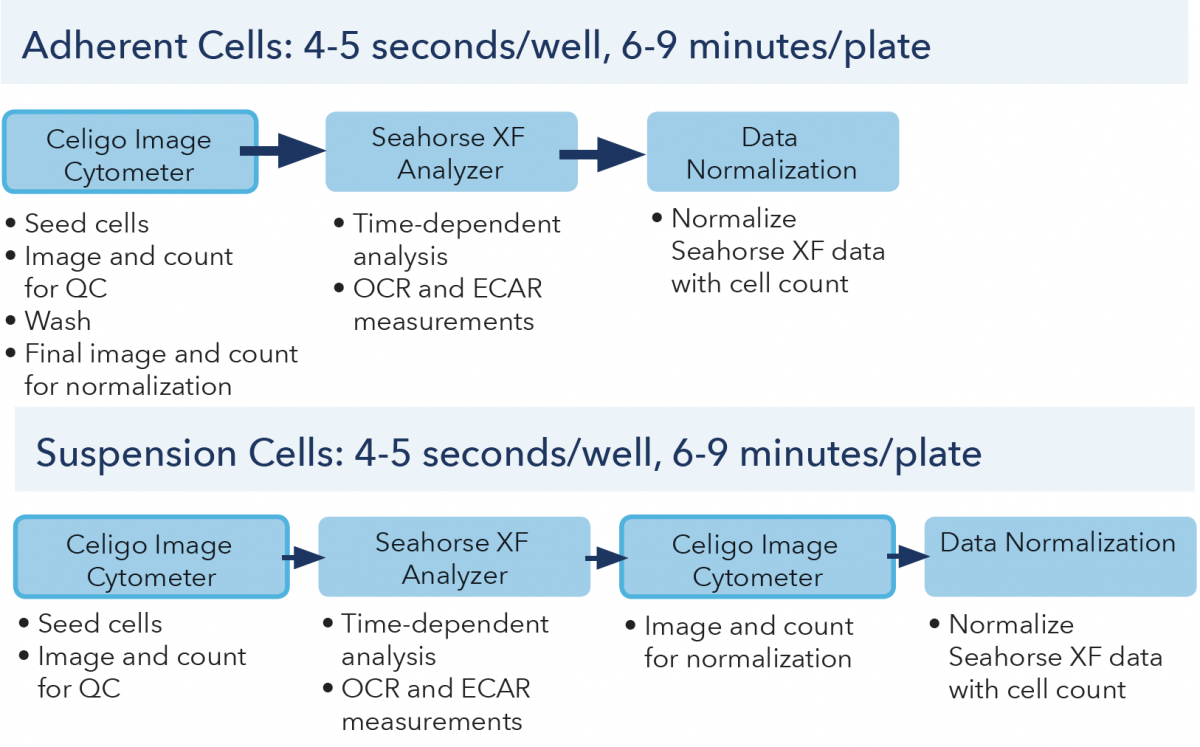
Figure 7. Celigo image cytometer workflows for adherent and suspension cells.
Whole well image and image quality
High image quality is an advantage of the Celigo image cytometer. Because of the proprietary F-Theta lens and Galvanometric Mirror, the Celigo instrument generates highly uniform and rapid whole well imaging in both brightfield and fluorescence for the entire plate. Therefore, it is unnecessary to extrapolate from one single image in the middle of the well, which may create uncertainty and inaccurate counting in the well if the cells are not uniformly seeded (Figure 8). These images can also help to identify issues with cells or incorrect seeding if assay results are not as expected.
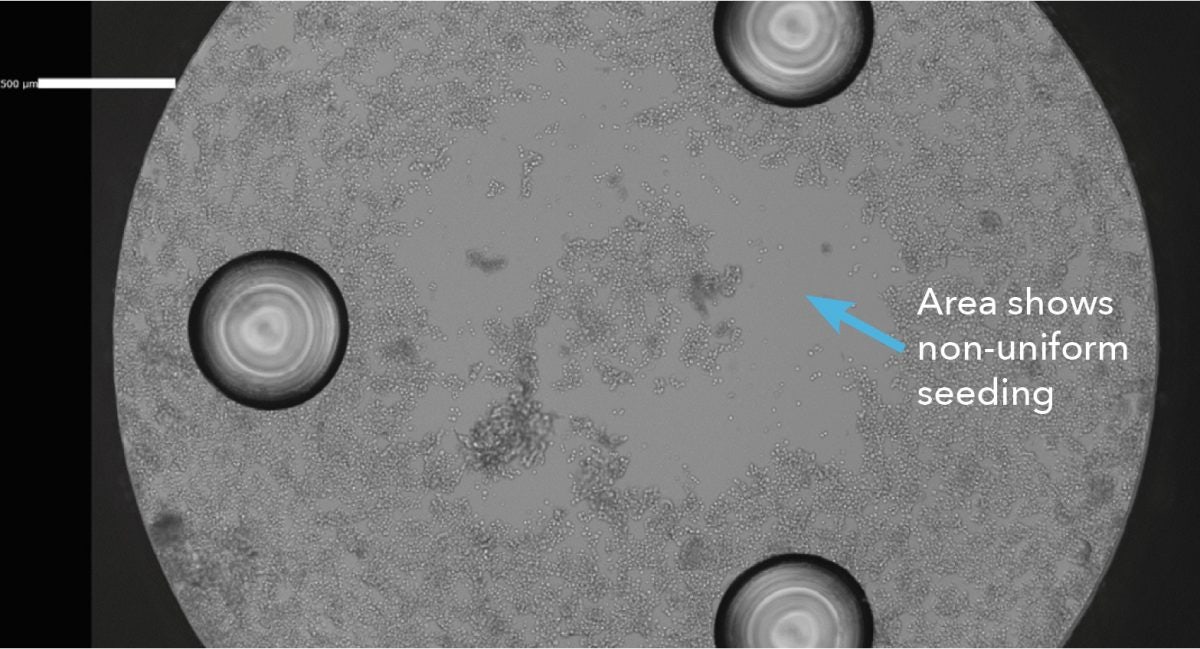
Figure 8. Celigo whole-well brightfield image shows non-uniformly seeded cells.
Sophisticated and reliable cell counting algorithm
Since cell shape and size is variable, researchers also require a flexible and sophisticated cell counting platform to ensure reliable results for normalization. Both adherent and suspension cells can utilize a variety but specific counting algorithm to count target cells in both brightfield and fluorescent images. In addition, location plots are used to specifically gate out the 3 post areas to further improve cell counting accuracy (Figure 2).
Perform label-free direct cell counting
The Celigo image cytometer can be used to directly count both adherent and suspension cells without fluorescent labels, which can remove the potential cytokinetic effect on the cells, as well as reducing the time for staining. However, since the Celigo system has one brightfield and 4 fluorescent channels (blue, green, red, and far red), fluorescence-based direct cell counting can also be easily performed.
References
- "Normalizing XF metabolic data to cellular or mitochondrial parameters", Application Note, Seahorse Bioscience
- "Normalization of Agilent Seahorse XF Data by In-situ Cell Counting Using a BioTek Cytation 5", Application, Seahorse Bioscience
- Riddle et al., "Expansion capacity of human muscle progenitor cells differs by age, sex, and metabolic fuel preference," Am J Physiol Cell Physiol, doi: 10.1152/ajpcell.00135, 2018
*Seahorse XF Analyzer is a product of Agilent.
For research use only. Not for use in diagnostic procedures.




























