
NEXTFLEX RiboNaut rRNA Depletion Kit (Human / Mouse / Rat)

NEXTFLEX RiboNaut rRNA Depletion Kit (Human / Mouse / Rat)




The NEXTFLEX® RiboNaut™ rRNA depletion kit (human / mouse / rat) is an effective method to remove rRNA contamination from mammalian total RNA while enabling labs to interrogate additional RNA species in a sample, not only limited to intact mRNAs.
| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | Ribodepletion |
The NEXTFLEX® RiboNaut™ rRNA depletion kit (human / mouse / rat) is an effective method to remove rRNA contamination from mammalian total RNA while enabling labs to interrogate additional RNA species in a sample, not only limited to intact mRNAs.


NEXTFLEX RiboNaut rRNA Depletion Kit (Human / Mouse / Rat)


NEXTFLEX RiboNaut rRNA Depletion Kit (Human / Mouse / Rat)


Product information
Overview
- Hybridization and bead-based protocol for rRNA depletion
- Optimized for use with 5 ng – 1 µg of total RNA as starting material
- Tested using the NEXTFLEX® Rapid Directional RNA-Seq Kit 2.0 and automated on the Sciclone® G3 NGSx and Zephyr® G3 NGS workstations
- Versions are available for automated and manual rRNA depletion
The NEXTFLEX RiboNaut™ Kit Ensures Effective rRNA Depletion using Hybridization Technology
The NEXTFLEX RiboNaut rRNA depletion kit (human / mouse / rat) utilizes subtractive hybridization technology consisting of a mixture of biotinylated oligos complementary to rRNAs to pull them out of the sample using streptavidin-coated magnetic beads. The kit has been optimized for use with 5 ng – 1 µg of total RNA as starting material to deplete cytoplasmic and mitochondrial rRNAs. It has been verified to be compatible with human, mouse, and rat RNA and it may be compatible with other mammalian species.
Additional product information

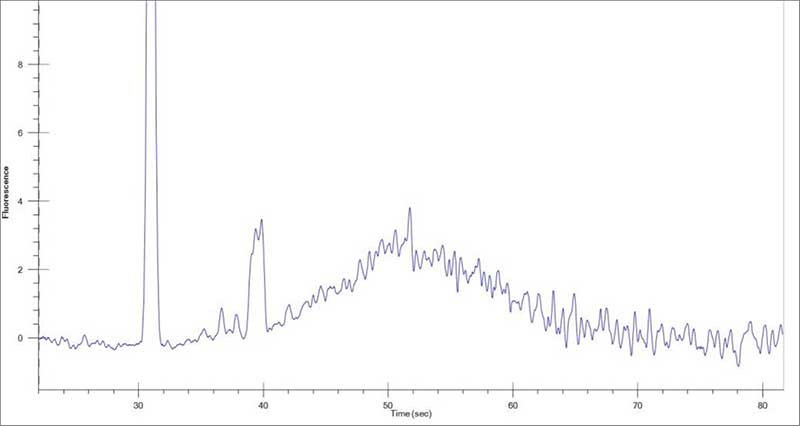
Figure 1: Example of 1 µg of Universal Human Reference RNA (Agilent® #740000) after rRNA depletion. 1 µL of rRNA-depleted RNA was run on the LabChip GXII Touch HT Nucleic Acid Analyzer using the RNA Pico Assay Reagent Kit (# CLS960012) and a DNA/RNA/Charge Variant Assay (# 760435).
Optimized to Produce Robust Performance Data from Total RNA Samples
The NEXTFLEX RiboNaut rRNA Depletion kit (human / mouse / rat) shows robust performance data using total RNA samples for gene coverage along a transcript, duplication rate, directionality, and rRNA contamination. Prior to rRNA depletion and subsequent RNA-seq, qualification of total RNA integrity using the LabChip GXII Touch HT Nucleic Acid Analyzer.
As an alternative mRNA-Seq solution for intact poly(A)-tailed mRNA species, Revvity offers the NEXTFLEX Poly(A) Beads 2.0 for use NEXTFLEX Rapid Directional RNA-Seq kit 2.0 or other applications.
Robust Gene Coverage
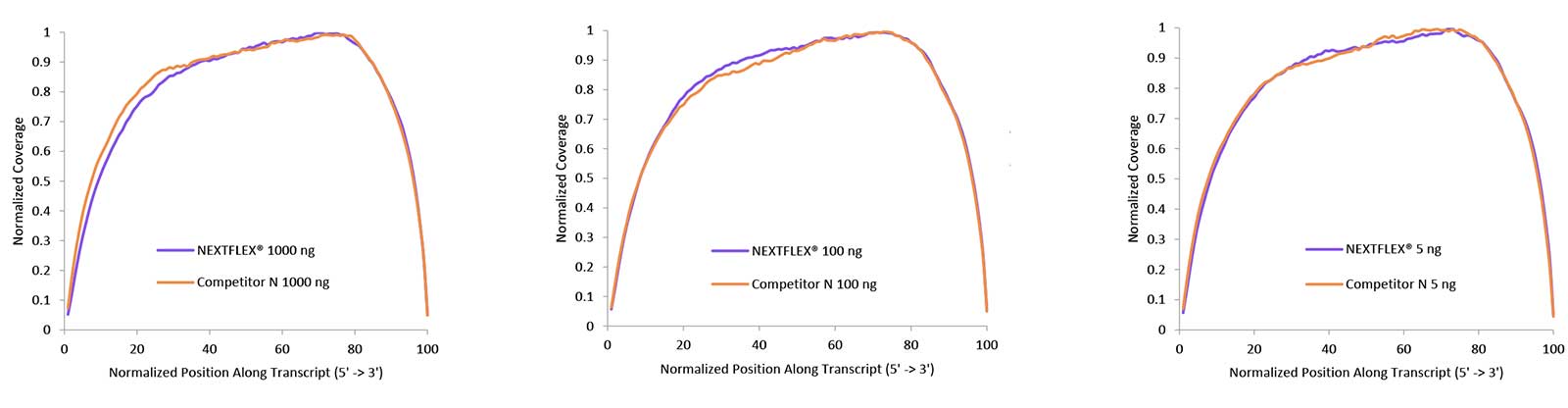
Figure 2. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrates even coverage along transcripts compared to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina® MiSeq® sequencer using single-end mode (1×151 bp). Reads were trimmed using cutadapt, mapped to the Gencode v30 reference transcriptome using bowtie2, and randomly downsampled to 720k reads. The coverage along transcripts was calculated using the BBMap pileup tool.
Low Duplication Rate
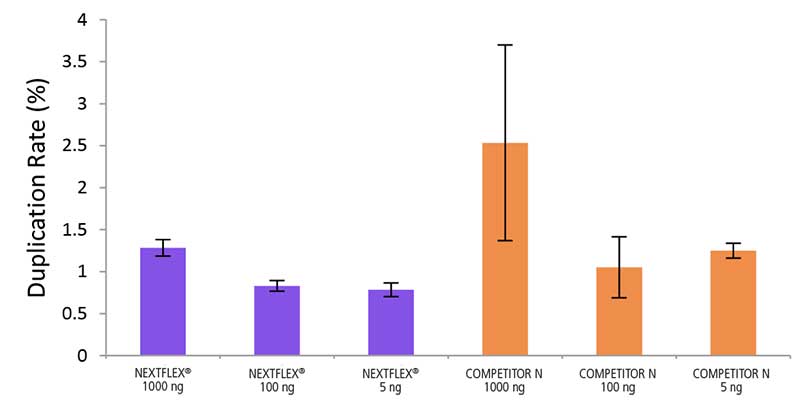
Figure 3. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrate low duplication rates compared to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using single-end mode (1×151 bp). Reads were trimmed using cutadapt, mapped to the Gencode v30 reference transcriptome using bowtie2, and randomly downsampled to 28k reads. Duplication rate was calculated using the fastp all-in-one FASTQ preprocessor.
Reliable Directionality
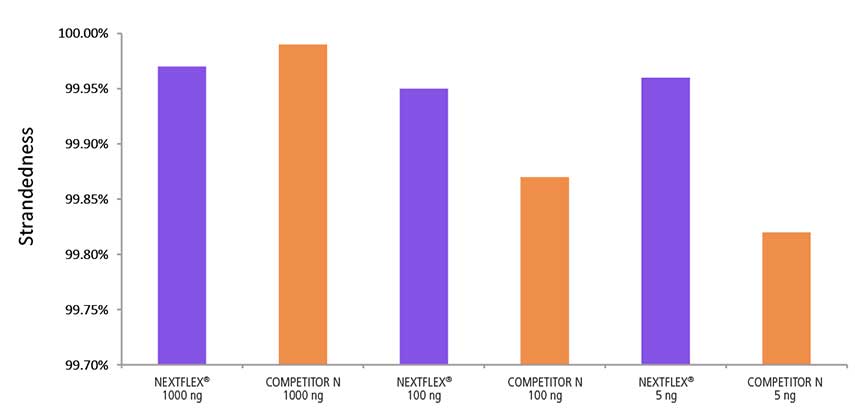
Figure 4. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrates comparable directionality relative to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using single-end mode (1×151 bp). Reads were trimmed using cutadapt and mapped to the Gencode v30 reference transcriptome using bowtie2. Reads from respective samples were combined and downsampled for a total of 800k reads each. Strandedness was calculated using the fastp all-in-one FASTQ preprocessor.
Low Levels of rRNA Contamination
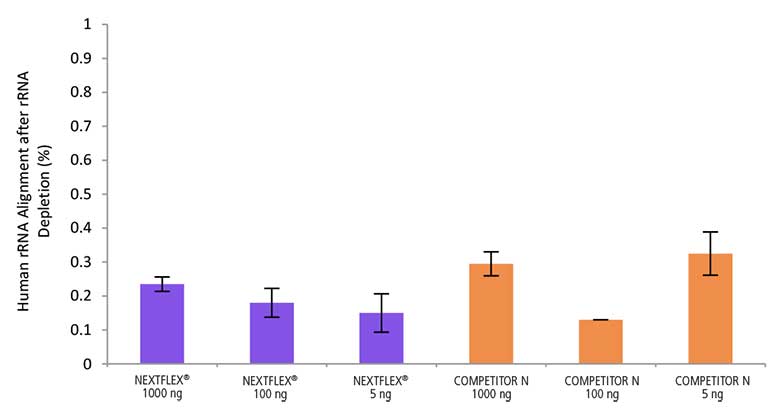
Figure 5. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 delivers libraries containing low levels of rRNA contamination compared to the Competitor N kit. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit (human, mouse, rat) and the Competitor N rRNA-depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0 and the Competitor N’s library preparation kit. The resulting libraries were sequenced on the Illumina MiSeq sequencer using single-end mode (1×151 bp). The reads were trimmed using cutadapt and the percent of rRNA was determined by using bowtie2 to map reads to human rRNA. The NEXTFLEX Rapid Directional RNA-Seq kit 2.0 demonstrated superior removal of 5S, 5.8S, 12S, 16S, 18S, and 28S rRNA species compared to the Competitor N kit.
Automated vs. Manual Prep
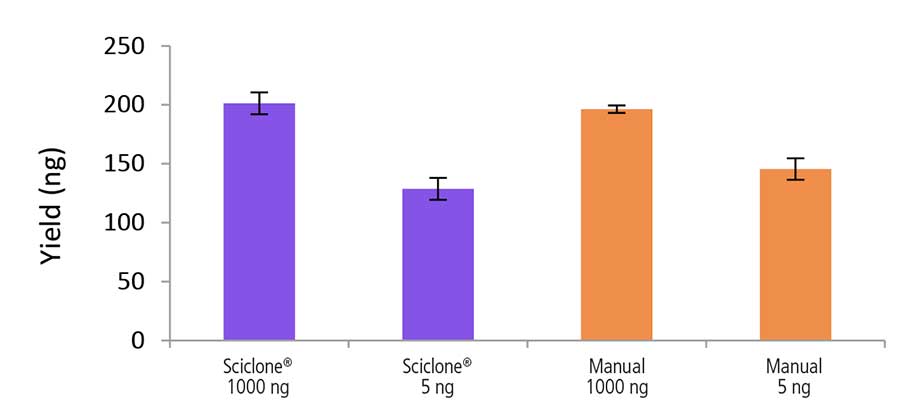
Figure 6. Libraries prepared using the Sciclone G3 NGSx workstation and manually deliver comparable yields using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0. rRNA-depleted total RNA was isolated from Universal Human Reference RNA (Agilent #740000) using the NEXTFLEX RiboNaut rRNA depletion kit. Libraries were generated using the NEXTFLEX Rapid Directional RNA-Seq kit 2.0. Final library concentrations were quantified using the Qubit ® 2.0 fluorometer (Thermo Fisher® Scientific #Q32866).
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Format |
Manual
|
| Product Group |
Ribodepletion
|
| Shipping Conditions |
Dual Temperature
|
| Unit Size |
8 rxns
|
Resources
Are you looking for resources, click on the resource type to explore further.


How can we help you?
We are here to answer your questions.






























