Directly measure viral transduction efficiencies by direct cell counting in 96-well plates
- No need to trypsinize cells and run on flow
- Perform whole-well imaging and analysis for an entire 96-well plate in <15 minutes
- Determine transient and stable transduction rates using live cell imaging and analysis
Introduction to optimization of cellular transfection and transduction
Optimization of cellular transfection and transduction includes choosing a protocol, determining the appropriate mass of plasmid/virus, and evaluating the optimum time after transfection/transduction for the best expression of the construct of interest. Using fluorescent reporters and non-destructive in situ imaging on the Celigo™ image cytometer, these parameters can be rapidly optimized by repeated imaging of one set of wells or flasks over a period of time. Whole-well brightfield cell counting generates cell-based data normalized to absolute cell numbers.
Protocol
- Plate 4 x 105 cells in a 6 well plate.
- Thaw virus vector stock at room temperature and store stock on ice
- Prepare a serial dilution ranging from 10-1 to 10-4
- Add 1 ml of diluted virus to pre-plated cells and incubate at 37°C
- Examine cells for GFP expression after 2 days of incubation
- Viral titer is determined by the percent of eGFP positive cells
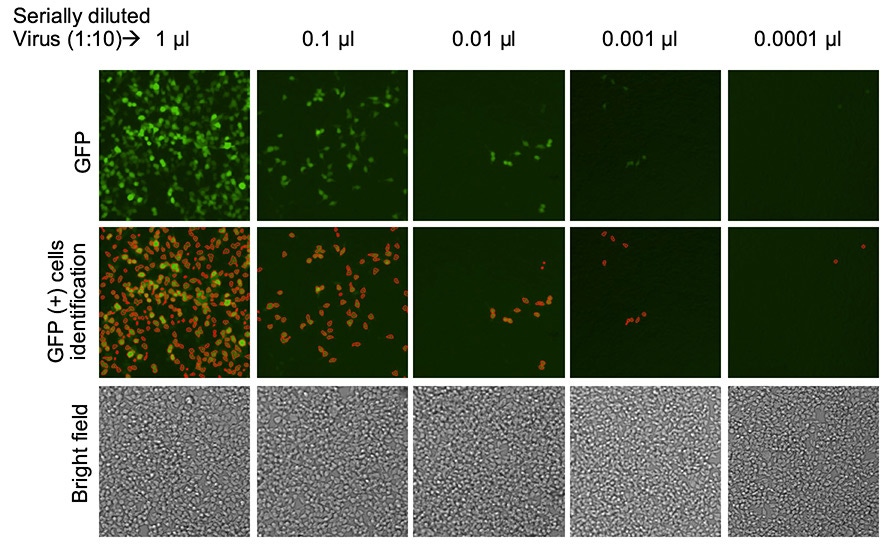
Fast and easy in-plate analysis of lentiviral transduction efficiency
- HEK293T cells were seeded and grown to 80 percent confluence
- Lentivirus was added to the wells at a 10-fold serial dilution
- After incubation, the Celigo was used to acquire brightfield images and GFP images for each well and analyze the data

Images show captured brightfield images for each dilution (bottom row), GFP positive cells for each dilution (top row) and Celigo software identified and counted (cells outlined in red) GFP positive cells (middle row).
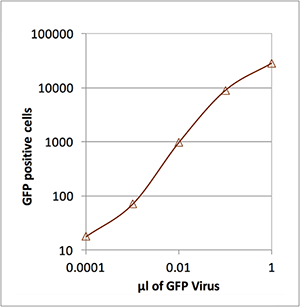
Number of lentivirus infected cells
| µl of GFP virus | # of GFP positive cells |
|---|---|
| 1 | 28,220 |
| 0.1 | 8,898 |
| 0.01 | 982 |
| 0.001 | 71 |
| 0.0001 | 18 |
This data shows that the Celigo has a wide range for cell counting. It counted as few as 18 GFP positive cells per well to over 28,000 GFP positive cells per well. The number of lentivirus-transduced cells was shown to be directly proportional to the amount of virus added to each well.
The Celigo live cell analysis of all labeled cells to determine the percent of cells expressing GFP in less than 15 minutes
By counterstaining the cells with Hoechst, the Celigo software identifies and counts each Hoechst positive and GFP positive cell. Because the Celigo counts every labeled cell in the well, it can automatically report the number of cells in each well as well as the percent transduction.
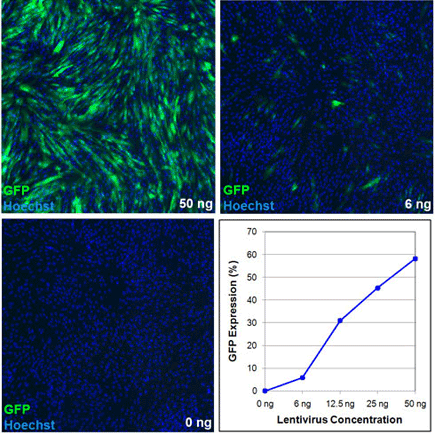
Images and analysis of transduced human foreskin fibroblasts
Images and image analysis of human foreskin fibroblasts transduced with varying amounts of a GFP lentivirus acquired and analyzed on the Celigo cytometer.
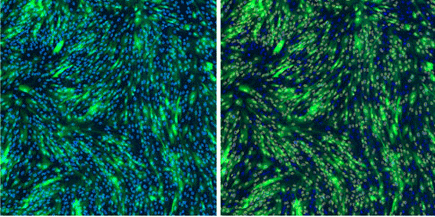
Live images: segmentation of transduced fibroblasts
Live images of segmentation using the Expression Analysis application. Nuclei are segmented as the mask (left) and GFP-positive nuclei are segmented as the target (right) to determine transduction efficiency.
For research use only. Not for use in diagnostic procedures.




























