Introduction to cell cycle
What is cell cycle?
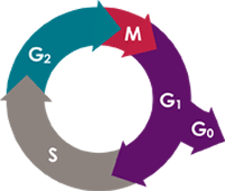
Cell cycle is defined as “the period between successive divisions of a cell.” Standard cell cycle of eukaryotic cells is divided into 4 phases.
Cell cycle phases

- G1 (G=Gap) phase. Cells are preparing for DNA replication. At this stage the cells contain about the same amount of DNA. The diploid number of a cell is the number of chromosomes in the cell. This number is commonly abbreviated as 2n, where n stands for the number of chromosomes. Since there are normally 23 pairs of chromosomes in human cells: 2n = 46
- S (S=Synthesis) phase. Replication of nuclear DNA occurs at this stage. During S phase there is a broad distribution of DNA content between G1 and G2 phase. The DNA content is doubled by the end of this phase. (2n to 4n)
- G2 (G=Gap) phase. Replication of the DNA is complete, and the cell is growing and preparing for division. At this stage, the cells contain twice the number of chromosomes (4n) that are found in G1.
- M (M=Mitosis) phase. Cell division. At the end of cell division each cell now contains the same amount of DNA: 2n
Importance of cell cycle research
Oncology
Since cancer cells often undergo abnormal cell division and proliferation, it is important to understand their cell cycle.
Pharmacology
Studies involving characterization of pharmacological reagents and their effect on cell cycle regulation.
Arrest normal cell cycling during chemotherapy treatment.
Cell/Molecular biology
To understand cellular, molecular, and biological mechanisms, it is essential to examine their role in cell cycle.
Current detection methods
Radioactivity measurement
3H-thymidine – Radioactive labeling of thymidine. Upon DNA replication, the radiolabeled thymidine is incorporated into new DNA.
Fluorescence-Based Detection
Live cell measurement
Hoechst 33342 – is cell permeable and therefore does not require cell fixation. Binds to the minor groove of double-stranded DNA.
Fixed cell measurement
Bromodeoxyuridine (BrdU)– a thymidine analogue, is incorporated into the genome during the S-phase of the cell cycle. Detected using anti-BrdU antibodies.
DAPI – binds to the AT rich regions of the DNA. Has poor live cell permeability and therefore often used post-fixing.
Propidium Iodide (PI) – A membrane exclusion dye and an intercalating agent that stains the cellular genome upon cell fixation.
Fluorescence-based, fixed cell measurement using cellometer
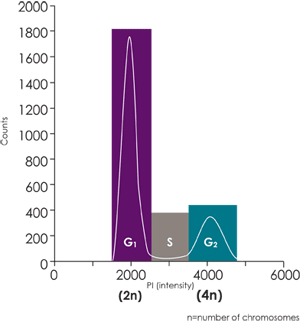
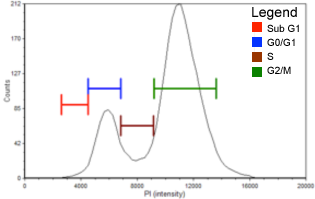
Labeling DNA with propidium iodide (PI) allows for fluorescence-based analysis of cell cycle.
This assay generates a cell population histogram (on right) in respect to PI fluorescent intensity.
Amount of PI fluorescent intensity is correlated to the amount of DNA within each cell.
Since the amount of DNA doubles (2n → 4n) between G1 and G2 phases the amount of fluorescent intensity of the cell population also doubles from 2000 to 4000 fluorescence units.
Cell cycle in 5 easy steps

1. Pipette 20 µl of sample into a disposable slide.

2. Insert slide into the cellometer instrument
3. Select assay from a drop down menu
4. Click count, acquire image and view cell count, concentration, and diameter
5. Export data to obtain cell cycle histogram results
Examples of cell cycle detection for cell lines
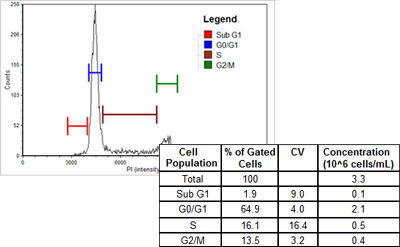
PC3
| Cell population | % of Gated cells | Concentration (10^6 cells/mL) |
|---|---|---|
| Total | 100 | 4.8 |
| Sub G1 | 0.1 | 0.0 |
| G0/G1 | 80.0 | 3.9 |
| S | 2.8 | 0.1 |
| G2/M | 16.2 | 0.8 |
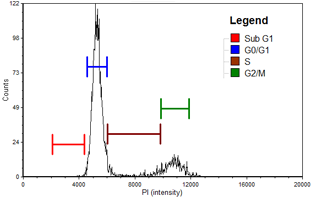
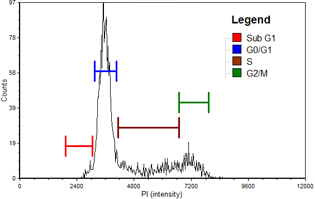
PANC-1
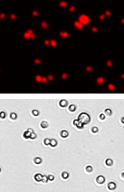
| Cell population | % of Gated cells | Concentration (10^6 cells/mL) |
|---|---|---|
| Total | 100 | 5.7 |
| Sub G1 | 1.2 | 0.1 |
| G0/G1 | 59.3 | 3.4 |
| S | 20.5 | 1.2 |
| G2/M | 18.4 | 1.1 |
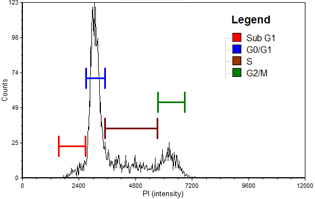
HeLa
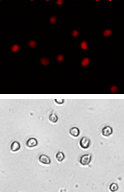
| Cell population | % of Gated cells | Concentration (10^6 cells/mL) |
|---|---|---|
| Total | 100 | 3.7 |
| Sub G1 | 2.2 | 0.1 |
| G0/G1 | 66.8 | 2.5 |
| S | 14.2 | 0.5 |
| G2/M | 14.6 | 0.5 |
Jurkat

| Cell population | % of Gated cells | Concentration (10^6 cells/mL) |
|---|---|---|
| Total | 100 | 5.2 |
| Sub G1 | 4.0 | 0.2 |
| G0/G1 | 62.3 | 3.3 |
| S | 16.4 | 0.9 |
| G2/M | 16.1 | 0.8 |
Cell cycle experiments
Protocol for fixed cell samples stained with PI
- Use the Cellometer Sample Adjustment Calculator to determine the sample volume required to obtain a starting number of 2 million cells.
- Spin down cell sample at 200 – 400 x g for 5 minutes then re-suspend cell pellet in 200 µl of PBS* (for a final concentration of 5 x 106 to 1 x 107 cells/ml.)
*If using methanol, do not re-suspend cell pellet in 200 µl of PBS. Re-suspend cell pellet in 500 µl of 100% ice cold methanol, and keep on ice for 15 minutes.
- Gradually add 500 µl of 200 proof (100%) ice cold ethanol to fix the cells. Gently mix by pipetting in and out at least ten times, and then keep on ice for 15 minutes. (Fixed cells may be frozen at -70°C for analysis at a later time.)
- Spin down cells sample at 200 – 400 x g (~1,000 to 2,000 rpm) for 8 minutes, remove PBS/Ethanol or methanol solution, re-suspend pellet in 150 µl of PI staining solution. Gently mix by pipetting in and out until there are no visible cell clumps in the sample (more than ten times).
- Incubate stained sample inside a 37°C incubator for 40 min.
- Spin down cells 200 – 400 x g (~1,000 – 2,000 rpm) for 8 minutes. Carefully, without disturbing the pellet, remove the staining solution. Then re-suspend the cells in 200 µl of PBS.
- Analyze stained cells within 30 minutes. Store in the dark prior to analysis.
Experimental data
Jurkat cells were used to analyze cell cycle kinetics following treatment with the cell-cycle-arresting drugs nocodazole and etoposide. Jurkat cells were incubated with media only (control), nocodazole (0.004, 0.02, 0.1 µg/mL), or etoposide (0.06 µM, 0.12 µM) for 24 hours. Control and drug-treated cells were fixed then stained with propidium iodide. For each sample, 20 µl of cell sample (at ~4 x 106 cells / mL) was loaded into a Cellometer counting chamber, inserted into the Vision CBA Analysis System*, and imaged in both brightfield and fluorescence. The fluorescence intensity for each cell was measured.
*The Vision CBA Analysis system has been superseded by the Cellometer Spectrum system.
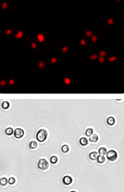
Brightfield image
The Cellometer instrument acquires a brightfield image for each sample tested. The bright field image allows researchers to verify cell morphology, evaluate the degree of homogeneity of the sample, and identify the presence of cellular debris.

Fluorescent image
Because all of the cells have been fixed, all of the cells are stained with propidium iodide and appear in the fluorescent image.
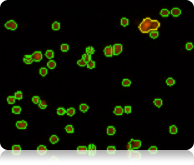
Fluorescent counted image
The fluorescent counted image can be used to confirm that cells are counted correctly. Individual counted cells are outlined in green. Uncounted cells are outlined in yellow. Cellometer software uses proprietary algorithms to accurately count individual cells within clumps.
Cell cycle histogram
A cell cycle histogram is automatically generated for each sample using the optimized cell cycle data layout in FCS Express™ Flow Software. Gating can be manually optimized directly on the histogram with automatic update to the associated data table. Histograms and data tables for control and experimental samples are shown in the experimental data section below.
Cell cycle analysis
For the control sample (Figure 1, Table 1), over half of the cell population is in the G0/G1 phase. There are also distinct populations of cells in the S phase (more than 20%) and the G2/M phase (more than 15%). The Sub-G1 group in the following histograms includes cellular debris and late apoptotic and necrotic cells. Gating was set for the control sample and applied to histograms for the two experimental conditions presented.
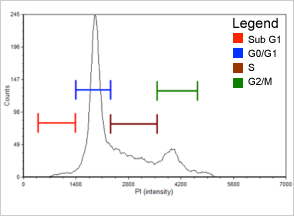
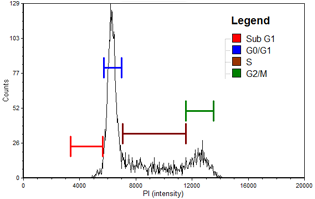
Figure 1. Control
Population histogram for the control sample generated with FCS Express™ software. Sub-G1 population is indicated in red, G0/G1 phase population is in blue, S phase population is in brown, and G2/M phase population is in green.
| Cell population | % of Total cells | Concentration (x 10^6 Cell/ml) |
|---|---|---|
| Total | 100.0 | 11.7 |
| Sub-G1 | 2.1 | 0.2 |
| G0/G1 | 54.0 | 6.3 |
| S | 26.4 | 3.1 |
| G2/M | 15.4 | 1.8 |
The addition of cell cycle-arresting drug etoposide, in a dose dependent manner, caused a shift in the cell population from the G0/G1 phase to the G2/M phase. (Figures 2 and 3).
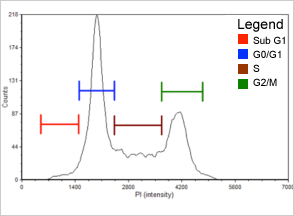
Figure 2. 0.06 µM Etoposide
Population histogram for the 0.06 µM etoposide sample generated with FCS Express™ software. There is a noticeable increase in the percent of cells that have arrested in the G2/M phase (see Table 2). Sub-G1 population is indicated in red. G0/G1 population is in blue, S phase population is in brown, and G2/M phase population is in green.
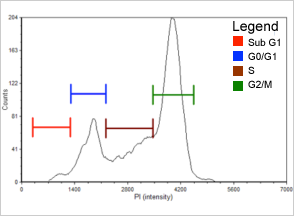
Figure 3. 0.12 µM Etoposide
Population histogram for the 0.12 µM etoposide sample generated with FCS Express™ software. At this drug concentration more than half of the cells are now arrested in the G2/M phase (see Table 2). Sub-G1population is in red, G0/G1 population is indicated in blue, S population is in brown, and G2/M population is in green.
| Cell population | % of Gated cells control (µM) | % of Gated cells etoposide (µM) | |
|---|---|---|---|
| 0.00 | 0.06 | 0.12 | |
| Sub-G1 | 2.1 | 2.8 | 2.2 |
| G0/G1 | 54.0 | 45.9 | 18.9 |
| S | 26.4 | 23.2 | 24.9 |
| G2/M | 15.4 | 26.8 | 52.2 |
Table 2 shows the population distribution for control and etoposide treated cells. As the etoposide concentration increased, the percent of cells that arrested in the G2/M phase of the cell cycle also increased from 15.4% to 52.2%
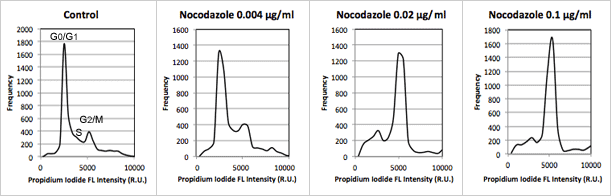
Figure 4
Population histograms for control, 0.004, 0.02, 0.1 µg/mL nocodazole samples generated with FCS Express™ software. Data for each phase of the cell cycle is show in the table on right. As the concentration of the nocodazole increased from 0.004 to 0.1 the percent of cells that arrested in the G2/M phase of the cell cycle increased from 12.3% to 47.7%
Table 3
| Cell population | Control (µg/ml) | Nocodazole (µg/ml) | ||
|---|---|---|---|---|
| 0.00 | 0.004 | 0.02 | 0.1 | |
| Sub-G1 | 6.9 | 6.7 | 12.1 | 9.1 |
| G0/G1 | 53.1 | 53.6 | 10.2 | 7.9 |
| S | 15.0 | 17.2 | 17.8 | 9.3 |
| G2/M | 14.3 | 12.3 | 41.4 | 47.7 |
Image cytometry correlation to flow cytometer
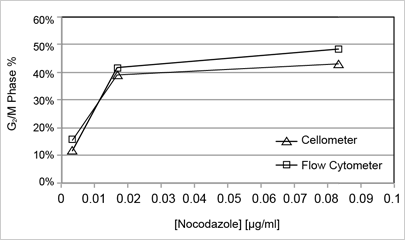
| Phase | 0.004 µg/ml | 0.02 µg/ml | 0.1 µg/ml |
|---|---|---|---|
| G2/M | 12.3 | 41.4 | 47.7 |
| G2/M | 15.2 | 39.1 | 43.1 |
The graph (right) shows the percentage of cells arrested at the G2/M phase following incubation with nocodazole. There was excellent correlation between the Cellometer Vision CBA Analysis System and the FACS-Calibur flow cytometer. Similar correlation data for etoposide is presented in the publication referenced below.
*The Cellometer Vision CBA system has been superseded by the Cellometer Spectrum.
Ultra Violet (UV) detection for cellcycle
There are a number of fluorescent-based dyes that are capable of binding to double stranded DNA. Along with PI (discussed above), DAPI and Hoechst are also two such dyes. Since the amount of bound fluorescent dye is directly proportional to the amount of DNA present within a cell, these dyes can be used to detect the cell cycle within a population of cells.
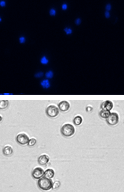
Below, is a representative example of cell cycle detection using DAPI. In this experiment, control and drug treated cells were stained with DAPI and analyzed using a Cellometer. Cells that were treated with 0.05 µg/ml of Nocodazole, arrested in the G2/M phase of the cell cycle compared to the control.
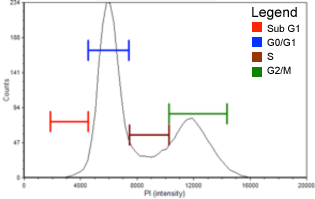
DAPI Control

| Cell population | % of Gated cells | Concentration (10^6 cells/mL) |
|---|---|---|
| Total | 100 | 2.0 |
| Sub G1 | 0.7 | 0.1 |
| G0/G1 | 55.9 | 1.1 |
| S | 10.2 | 0.2 |
| G2/M | 30.6 | 0.6 |
Nocodazole (0.05 µg/ml) treated
References
Cell cycle using cellometer
Chan, L., et al. (2011). Cellometer Vision as an Alternative to Flow Cytometry for Cell Cycle Analysis, Mitochondrial Potential, and Immunophenotyping. Cytometry Part A. 79A: 507-517.
Hana Schmeisser, et al. Type I interferons induce autophagy in certain human cancer cell lines. Autophagy, 9:5, May 2013.
For research use only. Not for use in diagnostic procedures.




























