
PG-Seq Rapid v2 kit

PG-Seq Rapid v2 kit




The PG-Seq™ Rapid kit v2 has been developed to analyze picogram quantities of DNA (single/multi-cells or low template DNA) from an embryo biopsy for preimplantation genetic testing. The PG-Seq™ Rapid kit v2 utilizes whole genome amplification (WGA) and next generation sequencing (NGS) technology to accurately screen all 24 chromosomes for whole chromosome aneuploidy and sub-chromosomal abnormalities. This PGT solution is compatible with Illumina® and Element Biosciences® sequencing platforms.
For research use only. Not for use in diagnostic procedures.
Revvity is a trademark of Revvity, Inc. All other trademarks are the property of their respective owners.
| Feature | Specification |
|---|---|
| Product Group | PGTA Library Prep |
The PG-Seq™ Rapid kit v2 has been developed to analyze picogram quantities of DNA (single/multi-cells or low template DNA) from an embryo biopsy for preimplantation genetic testing. The PG-Seq™ Rapid kit v2 utilizes whole genome amplification (WGA) and next generation sequencing (NGS) technology to accurately screen all 24 chromosomes for whole chromosome aneuploidy and sub-chromosomal abnormalities. This PGT solution is compatible with Illumina® and Element Biosciences® sequencing platforms.
For research use only. Not for use in diagnostic procedures.
Revvity is a trademark of Revvity, Inc. All other trademarks are the property of their respective owners.


PG-Seq Rapid v2 kit


PG-Seq Rapid v2 kit


Product information
Overview
Fast PGT-A prep: 3 hours from cell lysis to NGS ready
Updated, streamlined, simple PGT protocol
From DNA to data, the PG-Seq™ Rapid kit v2 includes all reagents required for cell lysis, whole genome amplification, indexing along with the PG-Find™ analysis software for automatic calling of aneuploidy and copy number variants used in PGS (preimplantation genetic screening).
Now you can schedule a one-on-one 30-minute online demonstration with our experts to cover everything you need to know about the PG-Seq™ Rapid protocol and data analysis with PG-Find. Please choose a timeframe that suits you best and we will provide you a Microsoft Teams personal link to join when the time comes. We look forward to speaking with you.
Accurate Copy Number Detection
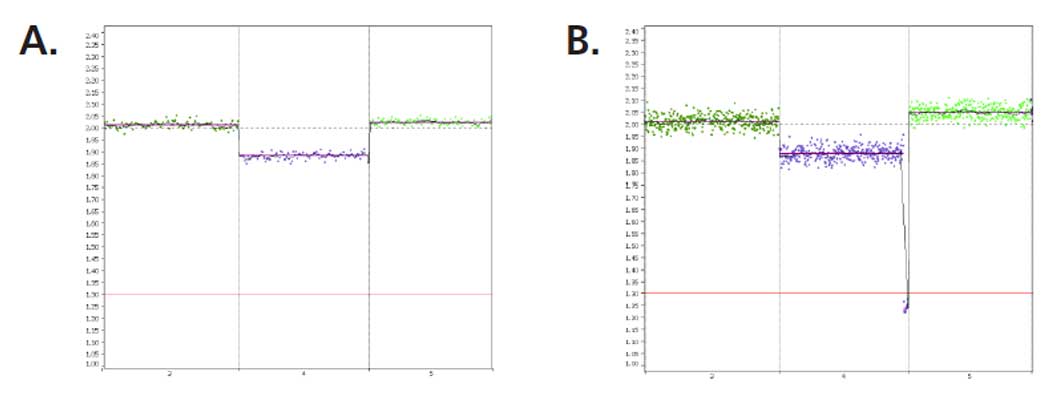
Extensively tested using over 100 cell lines and genomic DNA samples with known ploidy. Tested from whole chromosome aneuploidies down to segmental aberrations 7Mb in size with 30pg of genomic DNA or 5 cell fibroblast samples, representative of a trophectoderm biopsy. See PG-Seq™ Rapid kit v2 app note for more information.
Flexible Kit Format
The PG-Seq™ Rapid kit v2 has 48 rxns Two sets of 48 unique indexes are available, allowing up to 96 sample multiplexing (or up to 384 sample multiplexing available by custom request). Larger 40 µL WGA PCR 1 reaction volume enable options for alternative downstream processing. High WGA PCR 1 yield of 2-4 µg total DNA with only 23 cycles of PCR.
Improved whole genome coverage and accuracy
Enhanced PG-Find quality scores compared to previous versions of the kit signifies less noise, less bias, and higher confidence. Improved whole genome coverage when PG-Seq™ Rapid v2 WGA PCR 1 product is used in hybridization capture panels.
Additional product information
Sample Traceability in PGT-A
Sample traceability is crucial to ensure reliable embryo classification. This process involves tracking from the initial collection of the biopsy through to the final analysis.
For sample collection we recommend Cap2™ tubes, with 2D datamatrix code, free of DNAse, Rnase, human genomic DNA and compatible with automation.
To track samples individually we take advantage of the mitochondrial genome.
The mitochondrial genome is maternally derived and contains single nucleotide variants that can be used to distinguish between individuals. Revvity has shown that correct grouping of sibling embryos can be achieved in >98% of cases using mitochondrial DNA SNV profiling. The new mtDetect™ web app extracts mitochondrial DNA information from data obtained during the standard PG-Seq™ Rapid v2 workflow enabling the monitoring of external DNA contamination and the ability to detect sample swapping and mislabeled samples.
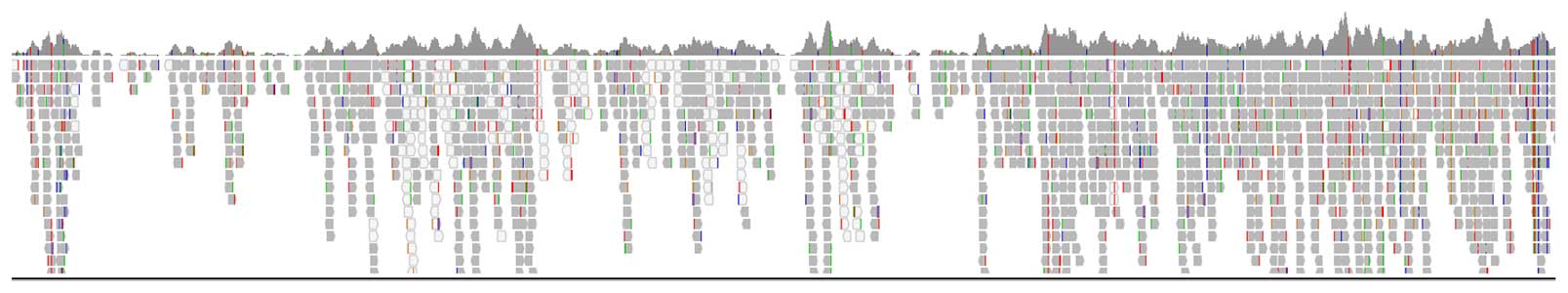
Figure 1 shows 90% coverage of the mtDNA genome using the PG-Seq Rapid v2 kit without any protocol modification. Embryo biopsies processed with the PG-Seq™ Rapid v2 kit analyzed with mtDetect™ web app can successfully be identified and grouped based on their mitochondrial DNA SNV profiles. Identity determination allows sample tracking and can assist in the identification and monitoring of possible external sample contamination along with mislabeled samples.

Figure 1: IGV coverage track of the full mitochondrial genome (chrM: 1-16364bp) from a PG-Seq Rapid v2 amplified 5-cell sample with 1,000,000 sequencing reads shows 90% coverage of the mtDNA genome.
Easy-to-Use, Highly Customizable Analysis Software
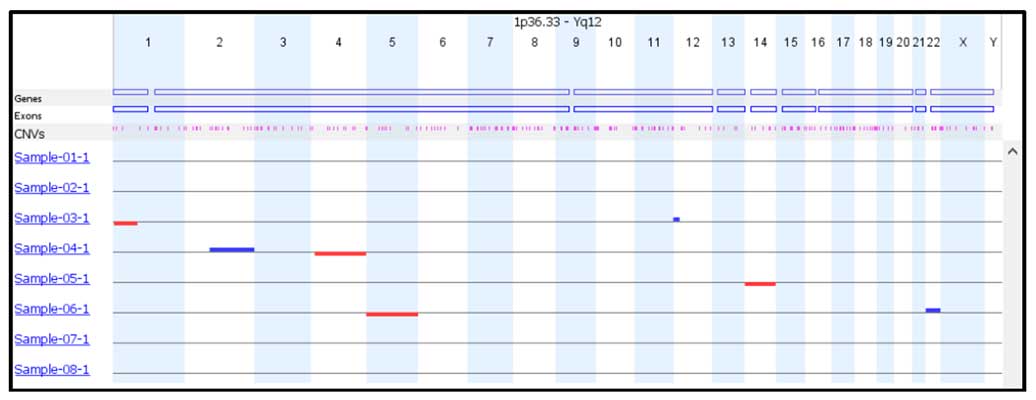
Two analysis options, self-reference and reference based, to accommodate every laboratory’s individual needs. Multiple visualization features including selecting which chromosomes to view and report, a single check box to view the raw vs smoothed data, unlimited color options, multiple result images. User adjustable event calling thresholds that can be used to designate mosaic, 1 copy or 2 copy gains and losses. An easy to interpret result overview screen allows the results of every sample to be easily analyzed and for general data trends to be observed.
A modifiable binning distance allowing the CNV resolution to be tailored according to laboratory requirements and sample sequencing depth and coverage.
Automatic and manual copy number calling of whole chromosome and sub-chromosome copy number events, including detailed information on the position and size of each event.
Specifications
| Product Group |
PGTA Library Prep
|
|---|---|
| Shipping Conditions |
Dual Temperature
|
| Unit Size |
48 rxns
|
References
- Avila Perez, C., Parnell, L., Florensa Bargallo, M., Herreros Cuesta, J., Larreategui Laiseca, Z., Prados Dodd, N., Ruiz Perez, M., & Wells, D. (2023). P-731: The accuracy of truly non-invasive PGT using spent culture media is insufficient to justify routine clinical use. Human Reproduction, 38(Supplement_1), dead093. 1050.
- Banu, M., Pathan, A. A. K., & Chaitanya, K. V. (2023). Diagnostics for Genetically Inherited Disorders: From Cytogenetics to Genomics Technologies-A Review. Biomedical and Pharmacology Journal, 16(2).
- Cheng, H. Y. H., Chow, J. F. C., Lam, K. K. W., Lai, S. F., Yeung, W. S. B., & Ng, E. H. Y. (2023). Randomised double-blind controlled trial of non-invasive preimplantation genetic testing for aneuploidy in in vitro fertilisation: a protocol paper. BMJ Open, 13(7), e072557.
- Curnow, E., Ryan, G. L., & Yu, B. (2023). Discordant non-invasive PGT-A results and clinical outcome. Fertility and Sterility, 120(4), e276.
- Spath, K., Costa-Borges, N., Nikitos, E., Kostaras, K., Calderón, G. C., Psathas, P., & Wells, D. (2023). P-730: Detection of mitochondrial reversal following meiotic spindle transfer: a finding of importance for mitochondrial replacement therapies used for the purpose of avoiding…. Human Reproduction, 38 (Supplement_1), dead093. 1049.
- Sonehara, H., Matsumoto, R., Nakayama, N., Kobanawa, M., Numata, K., Kawasaki, A., & Shozu, M. (2022). Aneuploidy and sex concordance rate between cell-free DNA analysis from spent culture media of preimplantation embryo and DNA from whole embryo with respect to different…. Reproductive Medicine and Biology, 21(1), e12493.
Resources
Are you looking for resources, click on the resource type to explore further.
In this application note we detail the performance and accuracy of the analysis software on embryos analysed with the PG-Seq™...


How can we help you?
We are here to answer your questions.






























